Rebecca Scheck
Tufts University
Talk Session: SESSION 1: POST-TRANSLATIONAL MODIFICATIONS
Date: Sunday, June 12, 2022
Talk Time: 09:35 am - 10:00 am
Talk Title: Learning the Rules for Selective Protein Glycation
Rebecca was born and raised just outside of NYC. As an undergraduate, she attended Columbia University and worked in the lab of Prof. Colin Nuckolls. For her Ph.D., she attended the University of California, Berkeley, where she worked with Prof. Matt Francis. She was then an NIH postdoctoral fellow with Prof. Alanna Schepartz at Yale University. Rebecca joined the Tufts faculty over the summer of 2013.
The Scheck lab focuses on the development of new chemical methodology that can be used to unravel complex post-translational modification, PTM, networks within a living cell. Our focus is on PTMs that have been particularly difficult to study using traditional tools, which typically inhibit or profile specific enzyme activities. In particular, we address this problem through the development of new, selective chemical methods that can predictably modulate and/or report on PTM function. These methods will be used to address longstanding questions about the role of vital, yet elusive, PTMs in inflammation, bacterial infection, autoimmune disease, cancer, as well as metabolic and age-related diseases.
The ongoing projects in our lab are divided into three main research areas, and are described in further detail below:
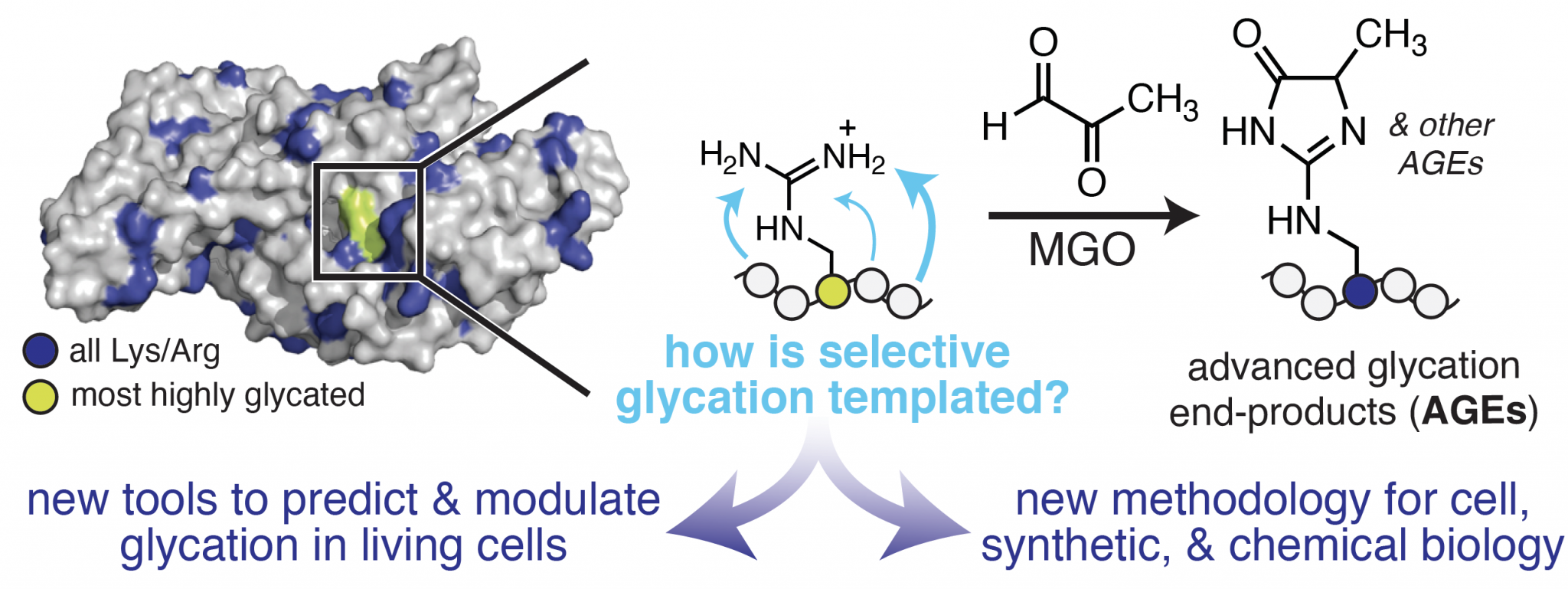
Uncovering the Chemistry & Biology of Protein Glycation
Our lab aims to develop a molecular understanding of the features that control protein glycation, a selective non-enzymatic PTM. Glycation is a hallmark of molecular aging associated with many diseases, including diabetes, cancer, Alzheimer’s disease, and age-related disorders of the skin and eye. Although past studies have demonstrated that glycation can impact protein activity and may influence disease development, we do not currently understand what controls the susceptibility of certain proteins to become glycated. This remains an open question preventing a molecular understanding of the biological role of glycation.
A major challenge that has hindered progress in this area stems from the fact that glycation occurs spontaneously and without an enzyme. This feature has rendered traditional chemical biology tools, which generally inhibit or profile specific enzyme activities, poorly suited for the study of glycation. Therefore, to evaluate the role of glycation as a functional PTM, new methods that can predict and control non-enzymatic chemistry in living cells are critically needed. Our lab is addressing this need using integrated chemical biology and mass spectrometry approaches that define the chemistry that governs selective glycation, and harness this information to develop a unique set of chemical tools that allow us to predictably modulate glycation outcomes in living cells.
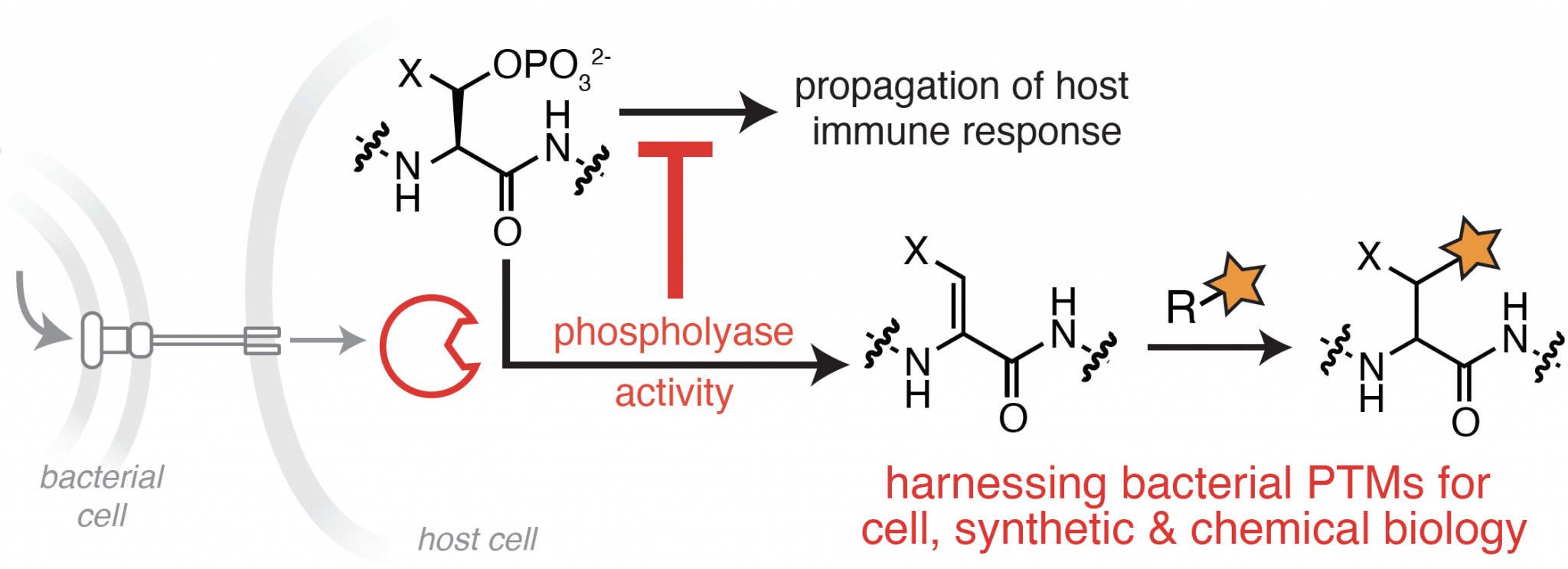
Exploiting the Unique Chemistry of Bacterial Enzymes
Bacterial effector proteins are key enablers of infection that act through diverse chemical interactions outside and within a host cell. Many effector proteins chemically modify host proteins to interrupt or rewire host signaling pathways, thereby promoting bacterial survival and replication. Among these, a subset of bacterial effectors promote transformations that do not occur naturally within the mammalian proteome, which we refer to as “orthogonal” PTMs, or oPTMS. Our lab is particularly interested in an oPTM catalyzed by bacterial phosphothreonine lyases, also referred to as phospholyases, which results in the elimination of phosphate from phosphorylated serine and threonine residues. Unlike phosphatases, which catalyze the reversible removal of phosphate through hydrolysis, phospholyases promote an irreversible elimination of phosphate that drastically disrupts endogenous signaling pathways.
Our work seeks to understand the full extent of selectivity within the phosphothreonine lyase family. This information is a crucial step towards identifying their complete repertoire of substrates and subsequent effects on host processes during infection. We are also harnessing this unique chemistry to identify new biomarkers for bacterial infection, and to develop new methods for proteome-wide detection of Ser/Thr phosphorylation events.
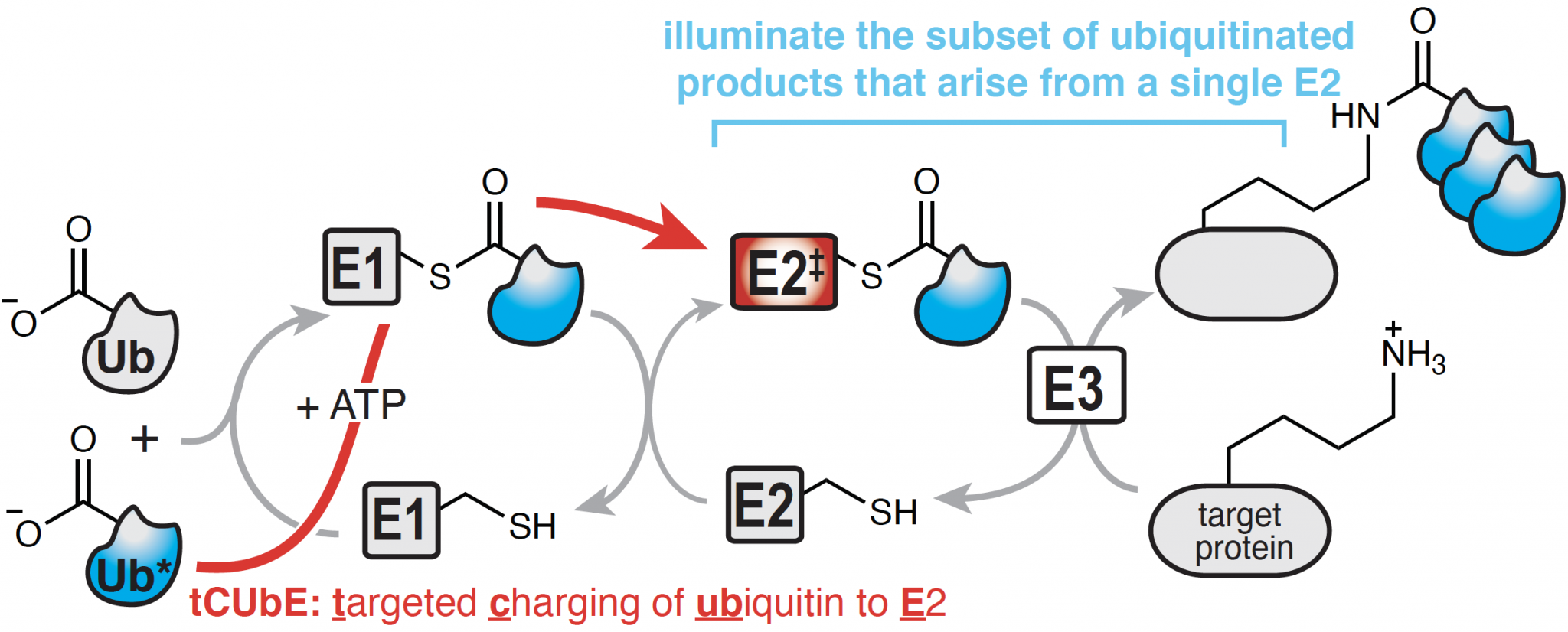
A Chemical Approach to Unravel the Ubiquitin-Proteasome System
The covalent attachment of ubiquitin to proteins is a vital posttranslational modification that controls regulated proteolysis and non-degradative signaling essential to all aspects of cellular function. When misregulated, perturbations within the ubiquitin-proteasome system, UPS can lead to many human pathologies including cancer. Ubiquitination relies on the precise transfer of a small protein, called ubiquitin, between three enzyme families, E1, E2, and E3, before becoming linked to its final protein target. The development of next-generation inhibitors that can selectively target specific steps within the vast UPS is an important frontier for cancer therapy. However, current therapies focus primarily on E3-substrate interactions, and current methods cannot unravel specific E1-E2-E3 sequences relevant for a particular protein target. Given that the UPS requires the coordination of hundreds of enzymes and thousands of specific interactions, it is extraordinarily difficult to study. As a result, there is a striking need for tools that can follow ubiquitin through the enzymatic E1-E2-E3 cascade to its target protein in live cells.
Our work in this area focuses on the development and implementation of such a method, called targeted Charging of Ubiquitin to E2, tCUbE. tCUbE channels a tagged ubiquitin, Ub*, to a specific E2 within a living cell, and can therefore identify distinct interactions relevant for ubiquitination of specific protein targets. This strategy enables us to track the fate of ubiquitin from a single member of the E2 family to a specific subset of E3 enzymes and/or target proteins with which the E2 interacts. As a result, this strategy is unmatched in its ability to illuminate the subset of Ub*-products that arise from the activity of a single E2, thus identifying new targets for disrupting the UPS for the treatment of cancer.
Research in the Scheck laboratory focuses on understanding protein post-translational modifications, PTMs, that have been difficult to study using traditional tools. Glycation is a PTM that occurs spontaneously and without an enzyme, yet it is known to occur selectively at certain sites on certain proteins.
We have uncovered molecular features that govern selective glycation, and we are now using this knowledge to develop new methods that predictably modulate glycation outcomes in living cells. This approach is uniquely suited to explore the biology of glycation by enabling the rigorous study of glycation as a functional PTM.